AWS News Blog
AWS Enables Consortium Science to Accelerate Discovery
My colleague Mia Champion is a scientist (check out her publications), an AWS Certified Solutions Architect, and an AWS Certified Developer. The time that she spent doing research on large-data datasets gave her an appreciation for the value of cloud computing in the bioinformatics space, which she summarizes and explains in the guest post below!
— Jeff;
Technological advances in scientific research continue to enable the collection of exponentially growing datasets that are also increasing in the complexity of their content. The global pace of innovation is now also fueled by the recent cloud-computing revolution, which provides researchers with a seemingly boundless scalable and agile infrastructure. Now, researchers can remove the hindrances of having to own and maintain their own sequencers, microscopes, compute clusters, and more. Using the cloud, scientists can easily store, manage, process and share datasets for millions of patient samples with gigabytes and more of data for each individual. As American physicist, John Bardeen once said: “Science is a collaborative effort. The combined results of several people working together is much more effective than could be that of an individual scientist working alone”.
Prioritizing Reproducible Innovation, Democratization, and Data Protection
Today, we have many individual researchers and organizations leveraging secure cloud enabled data sharing on an unprecedented scale and producing innovative, customized analytical solutions using the AWS cloud. But, can secure data sharing and analytics be done on such a collaborative scale as to revolutionize the way science is done across a domain of interest or even across discipline/s of science? Can building a cloud-enabled consortium of resources remove the analytical variability that leads to diminished reproducibility, which has long plagued the interpretability and impact of research discoveries? The answers to these questions are ‘yes’ and initiatives such as the Neuro Cloud Consortium, The Global Alliance for Genomics and Health (GA4GH), and The Sage Bionetworks Synapse platform, which powers many research consortiums including the DREAM challenges, are starting to put into practice model cloud-initiatives that will not only provide impactful discoveries in the areas of neuroscience, infectious disease, and cancer, but are also revolutionizing the way in which scientific research is done.
Bringing Crowd Developed Models, Algorithms, and Functions to the Data
Collaborative projects have traditionally allowed investigators to download datasets such as those used for comparative sequence analysis or for training a deep learning algorithm on medical imaging data. Investigators were then able to develop and execute their analysis using institutional clusters, local workstations, or even laptops:
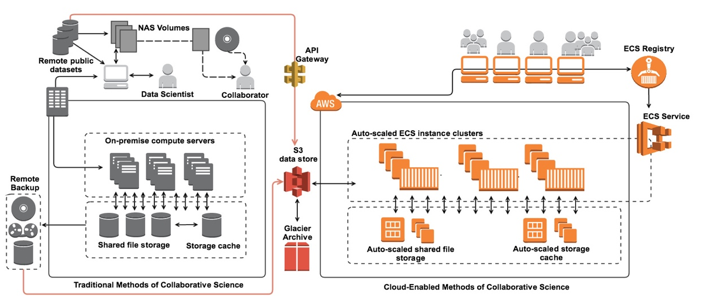
This method of collaboration is problematic for many reasons. The first concern is data security, since dataset download essentially permits “chain-data-sharing” with any number of recipients. Second, analytics done using compute environments that are not templated at some level introduces the risk of variable analytics that itself is not reproducible by a different investigator, or even the same investigator using a different compute environment. Third, the required data dump, processing, and then re-upload or distribution to the collaborative group is highly inefficient and dependent upon each individual’s networking and compute capabilities. Overall, traditional methods of scientific collaboration have introduced methods in which security is compromised and time to discovery is hampered.
Using the AWS cloud, collaborative researchers can share datasets easily and securely by taking advantage of Identity and Access Management (IAM) policy restrictions for user bucket access as well as S3 bucket policies or Access Control Lists (ACLs). To streamline analysis and ensure data security, many researchers are eliminating the necessity to download datasets entirely by leveraging resources that facilitate moving the analytics to the data source and/or taking advantage of remote API requests to access a shared database or data lake. One way our customers are accomplishing this is to leverage container based Docker technology to provide collaborators with a way to submit algorithms or models for execution on the system hosting the shared datasets:
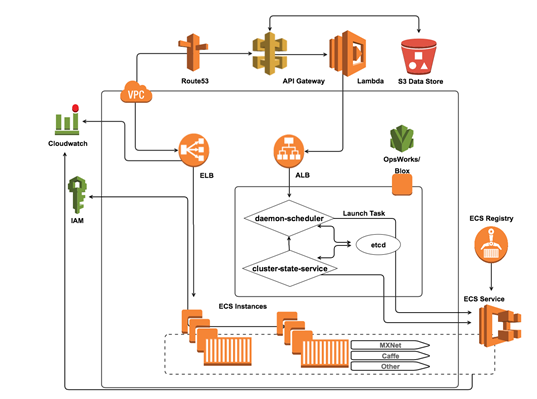
Docker container images have all of the application’s dependencies bundled together, and therefore provide a high degree of versatility and portability, which is a significant advantage over using other executable-based approaches. In the case of collaborative machine learning projects, each docker container will contain applications, language runtime, packages and libraries, as well as any of the more popular deep learning frameworks commonly used by researchers including: MXNet, Caffe, TensorFlow, and Theano.
A common feature in these frameworks is the ability to leverage a host machine’s Graphical Processing Units (GPUs) for significant acceleration of the matrix and vector operations involved in the machine learning computations. As such, researchers with these objectives can leverage EC2’s new P2 instance types in order to power execution of submitted machine learning models. In addition, GPUs can be mounted directly to containers using the NVIDIA Docker tool and appear at the system level as additional devices. By leveraging Amazon EC2 Container Service and the EC2 Container Registry, collaborators are able to execute analytical solutions submitted to the project repository by their colleagues in a reproducible fashion as well as continue to build on their existing environment. Researchers can also architect a continuous deployment pipeline to run their docker-enabled workflows.
In conclusion, emerging cloud-enabled consortium initiatives serve as models for the broader research community for how cloud-enabled community science can expedite discoveries in Precision Medicine while also providing a platform where data security and discovery reproducibility is inherent to the project execution.
— Mia D. Champion, Ph.D.